Community based study offers dental disease insights
August 3, 2023
From UNC Adams School of Dentistry
A multidisciplinary team of investigators at UNC-Chapel Hill, partnering with peers at University of Pennsylvania, published results from a community-based pediatric oral health study that found children’s microbiomes in the dental biofilm play major and previously unrecognized roles in dental disease. Their findings, reported in the journal Nature Communications, showed that dental caries, or tooth decay, in childhood is characterized by an imbalance in the oral microbiome and that certain previously unrecognized bacterial community members interact in complex ways in space and time.
The researchers, led by UNC Adams School of Dentistry’s Kimon Divaris, DDS, PhD, James Bawden Distinguished Professor, spent eight years studying the complex social, behavioral and biological determinants in children’s oral health. As part of this large NIH-funded effort (the ZOE 2.0 study), the investigators collected detailed clinical and biological information from 6,500 preschool-aged children enrolled in public preschools (i.e., Head Start programs) in North Carolina. The researchers collected saliva to study the children’s genomes, home water samples to measure fluoride concentration and dental plaque samples to assess the oral microbiome, all working toward a thorough study of oral health.
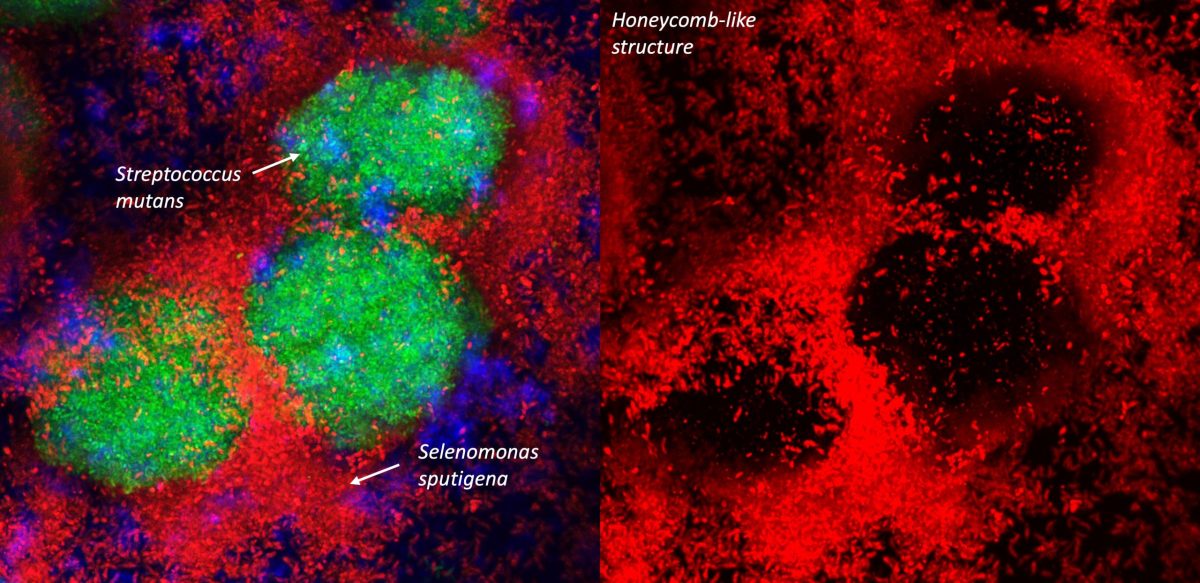
This graphic compares the bacteria Streptococcus mutans and Selenomonas sputigena, which were identified in the study.
“The paper reports the results of a very comprehensive study to try to understand microbial players in dental health and disease. We used data from a pilot sample of only 5% of our study participants, but the investigation was done in the most complete way it can be done,” said Divaris, the study’s primary investigator. “We began from a state-wide community-based study, collected detailed clinical data, social, behavioral and biological information and carried out state-of-the-art informatics and microbiome analyses.”
The UNC team of investigators also included Di Wu, PhD, associate professor of dentistry at the Adams School and associate professor of biostatistics at the UNC Gillings School of Global Public Health; Apoena De Aguiar Ribeiro, DDS, MS, PhD, associate professor at the Adams School; and Hunyong Cho, PhD, postdoctoral researcher in biostatistics at the Gillings School. They performed clinical, bioinformatics and laboratory studies using dental plaque samples from more than 400 child study participants. The team carried out whole genome sequencing and RNA sequencing analyses in partnership with UNC Microbiome Core Facility’s Andrea Azcarate-Peril, PhD, and Jeff Roach, PhD, then used custom databases and state-of-the-art informatics methods supported by Wu’s NIH-funded biostatistics and bioinformatics program to discover new caries-associated species.
“Microbiome data typically have excess zeros (about 70%) and high dimensions, meaning hundreds of species and millions of genes. We have evaluated 11 statistical models to deal with these challenges by generating comprehensive simulations,” Wu said. “The computational pipeline we developed ensures the strong candidate species, including Selenomonas sputigena, for downstream validation/mechanism study. Computational pathway analysis and correlation between species/genes provides further understanding of disease mechanisms and supports the wet lab experiment results.”
Subsequently, the team sought to replicate those findings using a second sample of children to see if the initial findings held true in an independent group of similarly-aged children.
“We were able to show that the abundance of at least 16 different species was associated with dental caries, and it’s not only their presence that’s important; it’s what they do and how they work together,” Divaris said.
In collaboration with a University of Pennsylvania team led by co-senior author Hyun (Michel) Koo, DDS, MS, PhD, further studies were designed to pinpoint the pathogenic role of these microorganisms. Koo and Zhi Ren, a postdoctoral fellow at Penn, developed a comprehensive series of biofilm studies involving computational imaging and animal experiments to validate the observational, clinical and bioinformatics findings. Using a novel, comprehensive multi-modal discovery-validation pipeline, the investigators then teased out how these oral bacteria might actually cause cavities. They identified an unexpected bacterial interaction and discovered a new contributor to tooth decay, Selenomonas sputigena. Their study’s results were published May 22 in Nature Communications and were highlighted in a recent NIH Research Matters story.
“It became clear that unless you employ additional virulence assessments as well as computational imaging and biogeography studies, it’s impossible to see how these species work together,” Divaris said. The combination of different methods, techniques, and expertise in this multidisciplinary team of investigators was arguably key to this discovery.
“It’s not only the presence of the bacteria that’s important but also what they do and how they work together,” Divaris said. “This unbiased, community-based study led to the discovery that species have the potential to cause dental disease in certain contexts, not only because of their ability to produce acid and demineralize enamel but because of their interaction in space with other species.” The investigators suggested that Selenomonas sputigena acts as a pathobiont in dental caries, with the implication that disrupting the relationship between these two species may offer a new strategy for preventing dental disease.
As the study expands into its next phase, Divaris said they hope to analyze already-collected plaque samples from the entire group of more than 6,500 study participants enrolled in the ZOE 2.0 study and follow up with them with a new clinical examination (the future ZOE 3.0 study), monitoring for incidence of disease and identifying additional oral health and disease biomarkers.
“The biggest success for us is the collaborative nature of this study. We had the patience to sit on data we generated four years ago until we could establish new collaborations, involve additional experts, and allow other team members to weigh in and do experimental work to add to what we were seeing,” Divaris said. “We are thankful to our study participants, funders, and U Penn collaborators. We’re all very satisfied. It was a great example of a community-based study powering biological discovery, and it’s very satisfying to see this scientific pipeline in motion. We saw it through the whole process, and that’s rare to see in dentistry. We hope to employ this multi-modal discovery-validation pipeline again in the near future.”
Contact the UNC Gillings School of Global Public Health communications team at sphcomm@unc.edu.
