UNC research identifies genetic variations linked to brain activity, mental health
May 27, 2022
Research from the Biostatistics and Imaging Genomics analysis lab – Statistics & Signal (BIG-S2) at UNC-Chapel Hill has identified 45 genetic variations in the brain that could contribute to development the development of disorders like depression, schizophrenia and Alzheimer’s disease.
These common but previously undiscovered variations could help health care providers better understand the genetic factors that impact behavioral and mental health and identify those who may be at higher risk for neurological disorders.
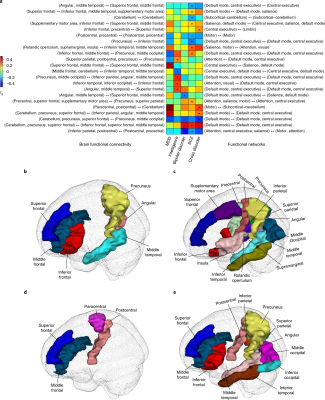
Published recently in Nature Genetics, the research team used resting state functional magnetic resonance imaging (rsfMRI) from more than 47,000 people to discover and validate common genetic variants that can influence brain activity. Of the 45 genetic regions they discovered that were linked with brain function, many had associations to the central executive, default mode and salience networks involved in the triple-network model of psychopathology. This model is the foundation for research on a range of mental disorders, including dementia, schizophrenia, depression and autism.

Dr. Bingxin Zhao

Dr. Tengfei Li
The study was a collaboration between Purdue University, the UNC Gillings School of Global Public Health and the UNC School of Medicine. It was co-authored by Gillings School alumni Bingxin Zhao, PhD, assistant professor at Purdue University, and Tengfei Li, PhD, in the UNC Department of Radiology. Other authors included biostatistics doctoral students Yue Yang, Di Xiong, Xifeng Wang, Tianyou Luo, Ziliang Zhu, and Professors Yun Li, PhD, and Hongtu Zhu, PhD. Zhu served as the corresponding author of this study. Faculty from other universities — such as Stephen M. Smith, PhD, from the University of Oxford — also contributed to this study.
The research team identified genetic elements that impact brain functional connectivity between each pair of 76 independent components and located numerous associated genes. In a perspective piece accompanying the study, the authors suggest that it “provides a step forward in understanding the genetic architecture of brain functional networks and their genetic links to brain-related complex traits and disorders.”
The team used the Genome-Wide Association Studies (GWAS) technique for 1,777 intrinsic brain activity traits and 9,026,427 common variants in a British data sample and validated by additional independent datasets. They identified 241 lead independent variants and discovered 603 significant locus–trait associations with 191 traits in 45 genomic regions.
The team reported that functional connectivity within the triple network had significantly higher heritability than those outside the triple networks, indicating a higher level of genetic control.
The researchers also examined genetic correlations between 484 heritable intrinsic brain activity traits to find if genetically mediated brain structural changes were associated with brain function. The results were linked to variants for a wide range of traits, including psychiatric disorders (such as schizophrenia, major depressive disorder, cross disorder and bipolar disorder), smoking and drinking, and brain structure and anthropometric traits. This led them to discover that intrinsic brain function has wide genetic links to many brain-related complex traits and clinical outcomes. This could contribute to future research that identifies brain functional mechanisms indicating a higher risk for these disorders.
“Until now, the relationship between genetic variants and intrinsic brain discovery has remained largely undiscovered,” the research team said. In the future, they expect that “accumulating publicly available imaging genetics data resources and more powerful feature extraction pipelines will lead to a better understanding of specific genes involved in human brain structure-function relationships.”
For more results on imaging genetics and information on future research, please visit the team’s Brain Imaging Genetics Knowledge Portal.
Contact the UNC Gillings School of Global Public Health communications team at sphcomm@unc.edu.