Data Science at the Gillings School
Biostatistics is the core of data science and its application to human health.
As massive, complex and varied data sets are produced through biomedical research, the necessity of skills possessed by biostatisticians is more evident than ever. Biostatisticians are best suited to the design, generation, and analysis of data. They possess the knowledge and tools needed to ensure the application of statistics to data science and human health is both factual and cutting edge, with reproducible results. The Department of Biostatistics at the UNC Gillings School of Global Public Health is charged with training and equipping future leaders in biostatistics and data science who will be integral to improving human health around the globe.
How are we involved in data science?
Genomic Data Science
Many faculty in the department work at the intersection of Genomics and Data Science. Research programs for these faculty are often highly collaborative, involving other members of the UNC community, including from the School of Medicine, School of Dentistry, as well as other departments within Gillings. Data Science projects with genetic or genomic data often involve tackling challenges such as technical biases, data sparsity or missingness, and effective integration of biologically relevant signal across multiple data modalities. Due to the high dimensionality and heterogeneity of genomic data sets, this work often taps into the latest developments in machine learning and causal inference. A key aspect of the research of faculty and students in the Department of Biostatistics in Genomic Data Science is the development of reproducible and transparent computational workflows, including the dissemination of new methods and algorithms as open source software.
NHLBI Trans-Omics for Precision Medicine Whole Genome Sequencing Program
Yun Li, PhD, associate professor of biostatistics and genetics, is a project investigator for the Whole Genome Sequencing (WGS) project of the Trans-Omics for Precision Medicine (TOPMed) program sponsored by the National Heart, Lung and Blood Institute. To support the NHLBI TOPMed program, the WGS project applies whole-genome sequencing to tens of thousands of previous NHLBI-funded study participants with well characterized phenotypes and existing clinical outcome data, as well as additional non-genomic omics profiles. This project aims to identify genetic markers of increased or decreased risk of heart, lung, blood and sleep (HLBS) diseases, as well as those that help define disease subtypes.
Li and her colleagues employ their expertise in statistics and data science to compile, analyze and make inferences about data on a massive scale. Ultimately their objective is to establish a novel genomic resource that is reflective of the diversity of the US population.
Learn more about TOPMed and WGS.
Causal Inference Research Laboratory (CIRL)
Biostatistics professor Michael Hudgens, PhD and epidemiology professor Stephen R. Cole, PhD were awarded one of four Gillings Innovation Labs administered by the Gillings School’s Research and Innovation Solutions unit in 2016. Cole, Hudgens and their team work to address two significant topics in biostatistics and epidemiology, namely “big data” analysis and causal inference, and will apply the methods to HIV and renal disease.
The CIRL deals with such Big Data as is collected by the Centers for AIDS Research (CFAR) Network of Integrated Clinical Systems (CNICS), which was developed to support population-based HIV research in the U.S. The CNICS cohort includes more than 30,000 HIV-positive adults engaged in clinical care from Jan. 1, 1995 to the present at eight CFAR sites across the nation. Electronic health records provide data on clinical events, laboratory measurements, and antiretroviral medications.
A Bayesian Spatial Model for Predicting the Location of Head Impacts
The location of a head impact is of particular interest for studying the biomechanics of head impacts, and may be useful for diagnosing and treating disease in the future. An accelerometer device is placed in a football helmet and outputs the direction (unit vector in 3D-space) and magnitude (scalar) of an impact’s peak linear acceleration (PLA). However, the magnitude and direction come from different vectors. As such, the two cannot be used as a single object and the direction on the unit vector scale must be predicted.
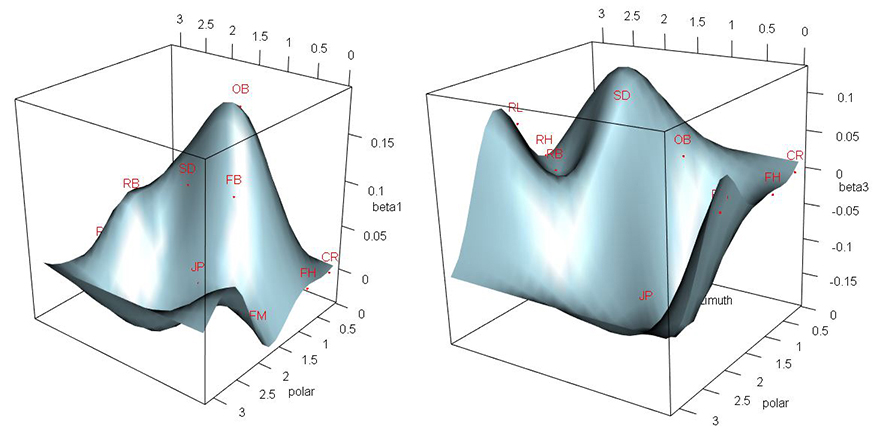
These two graphs—regression surfaces for the model’s parameters—show researchers a linear dependency between accelerometer device’s output and the output of a head form that records head impact data. In the graphs, it is the departures of the betas from zero that show this linear dependency. Otherwise, a flat plane would be the result.
Unfortunately, experimental data has shown accelerometer devices give imperfect output. As a result their utility as on-field clinical tools is limited and the validity of the data they produce for analysis is compromised. In order to correct the output of accelerometer devices and predict the true location of head impacts, UNC-Chapel Hill researchers developed a Bayesian spatial model based on the projected normal distribution, with spatial smoothing of model parameters built in by means of Gaussian process priors. The researchers assessed the model’s performance through simulation and applied it to data from a recent football helmet study. They showed spatial dependencies improve the model’s flexibility by comparing it to a model without those dependencies.