SRP researchers identify two new genera of bacteria associated with PAH degradation in soil

Dr. Michael Aitken

Dr. David Singleton
These microbes represent specific strains within larger groups of previously uncharacterized bacteria described by the research team as “Pyrene Group 1 (PG1)” and “Pyrene Group 2 (PG2)”, so named due to their initial association with the PAH pyrene. Recently, researchers were able to isolate and describe two new genera of bacteria within these groups.
PG1 is now represented by Rugosibacter aromaticivorans and PG2 by Immundisolibacter cernigliae, named after Dr. Carl Cerniglia, a former member of the UNC SRP External Advisory Committee and renowned microbiologist who has contributed greatly to understanding the biology of PAH-degrading bacteria. The Immundisolibacter strain is so different from described bacteria that researchers also proposed a new family and order to contain it. Researchers also discovered that these strains can degrade more compounds than just pyrene, with Immundisolibacter in particular capable of degrading a wide range of PAHs.
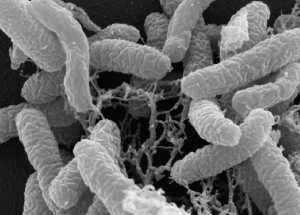
Rugosibacter aromaticivorans grown on pyrene
PAHs are formed during the incomplete burning of coal, oil and gas, garbage, or other organic substances like tobacco or charbroiled meat. They are usually found as part of a complex mixture in contamination sources such as coal tar. PAHs are found at many hazardous waste sites, especially former manufactured gas plants (MGPs) and facilities that treated wood with creosote. Some are considered to be carcinogenic, posing a significant threat to human and environmental health.
This discovery builds off of previous research in Project 5 (led by Dr. Aitken), which resulted in the detection of groups of bacteria previously unassociated with the degradation of PAHs in contaminated soil. The research team used a method called stable isotope probing (SIP) to originally detect these microbes.
This method uses PAHs labeled with heavier-than-normal carbon atoms (carbon-13). When bacteria living in contaminated soil consume these PAHs, they take in these heavier carbons, and use them to build new cells, including new, heavier, carbon-13-labeled DNA. Then, when the DNA is extracted from soil bacteria, it can be separated based on density and the bacteria which consumed the labeled PAHs identified.
However actually obtaining specimens that matched DNA sequences obtained by SIP proved more challenging. “Even after the organisms were first identified by SIP, it took many years before they could be isolated as pure cultures. The key was the development of a culture medium developed by Dr. Singleton that contained all of the necessary ingredients for these organisms to grow,” explained Dr. Aitken.
“Less than one percent of all environmental bacteria have ever been grown in the lab, for a variety of reasons, known and unknown,” added Dr. Singleton. “There was a fair amount of luck involved in finding the specific conditions under which these specific bacteria would grow.”
Collaborations with both the Chemistry Core and the Biostatistics Core were essential to identifying and characterizing these new bacterial strains. The Chemistry Core synthesized a number of uniformly labeled PAHs necessary for stable isotope probing, including naphthalene, phenanthrene, anthracene, fluoranthene, pyrene, and benz[a]anthracene.
Collaborations with the Biostatistics Core also enabled the research team to obtain complete genome sequences for these bacteria. Interestingly, DNA sequences from both of these genera appear frequently in gene libraries of PAH-contaminated soils around the world. For example, genes highly similar to those in the Immundisolibacter strain are also abundant in the creosote-contaminated soil that was brought from Spain by Dr. Joaquim Vila, a Marie Curie Research Fellow working in the Aitken lab. Researchers hope that the recent publication of isolation methods used to identify these bacteria will lead to new strains being identified and described.
Related research from Project 5 on using surfactants to enhance bioremediation of PAHs is the subject of the February 2017 NIEHS SRP Research Brief.
Relevant Citations:
Corteselli EM, Aitken MD, Singleton DR. (2016). Rugosibacter aromaticivorans gen. nov., sp. nov., a novel bacterium within the family Rhodocyclaceae isolated from contaminated soil, capable of degrading aromatic compounds. Int J Syst Evol Microbiol. doi: 10.1099/ijsem.0.001622
Corteselli EM, Aitken MD, Singleton DR. (2016). Description of Immundisolibacter cernigliae gen. nov., sp. nov., a high-molecular-weight polycyclic aromatic hydrocarbon-degrading bacterium within the class Gammaproteobacteria, and proposal of Immundisolibacterales ord. nov. and Immundisolibacteraceae fam. nov. Int J Syst Evol Microbiol. doi: 10.1099/ijsem.0.001714. [Epub ahead of print]
